
There are also many stray points, which are likely just noise.
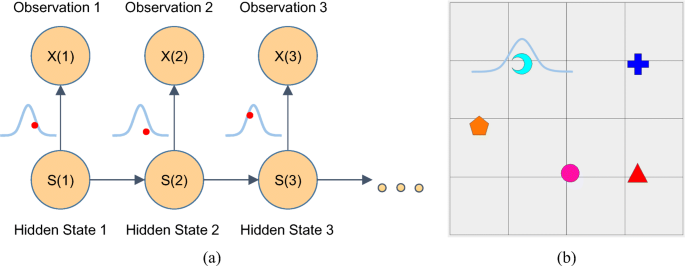
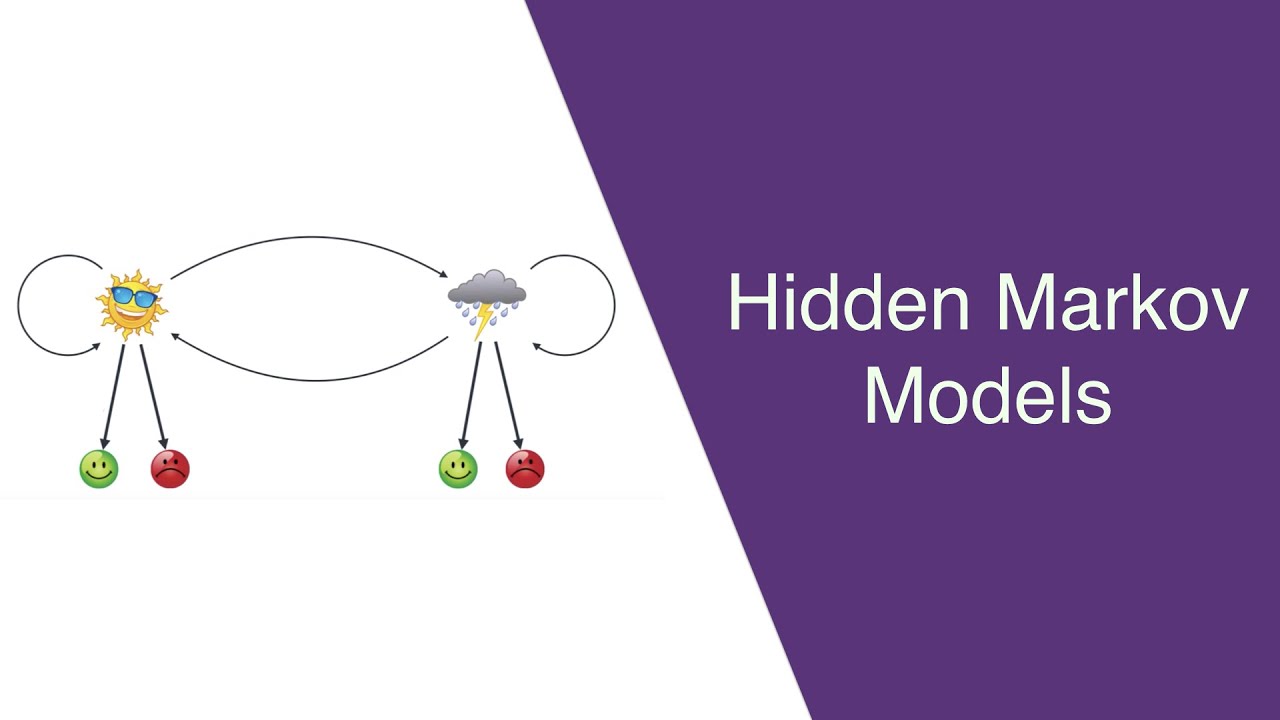
One can clearly see two amplifications and one deletion by eye. If we draw all the values, lined up along the genome, it looks like this: library(DNAcopy) After selecting only the mapped data from chromosomes 1-22 and X, there are 2271 data points. In particular, we chose the studies GM05296 and GM13330. The data correspond to two array CGH studies of fibroblast cell strains. We selected a subset of the data set presented in Snijders et al. (2001). If we have time we will also cover this algorithm.) (Aside: DNAcopy implements another useful signal processing algorithm, called circular binary segmentation. We load an arra圜GH dataset from a Bioconductor package, DNAcopy. Glossing over the technological details, if the signal is around 0 it means the sample has the same number of copies of DNA as the reference consistent stretches of higher or lower values than 0 indicate amplifications and deletions, respectively. Array comparative genomic hybridization (arra圜GH) is a microarray technology which compares, at a set of regions tiling the genome, the amount of DNA from a sample compared to a reference. Here we will present the Hidden Markov Model (HMM), and apply it to detect copy number variants in arra圜GH data. Hidden Markov Models and Dynamic Programming Michael Love


 0 kommentar(er)
0 kommentar(er)
